Without Antibodies and Without Amplification: Ultra-fast Identification of Whole Proteins Using a Technology Developed at the Technion
Researchers envision a future where a single blood test can deliver rapid, precise protein diagnostics — revolutionizing cancer care and advancing protein science
A groundbreaking technology developed in the laboratory of Prof. Amit Meller from the Technion – Israel Institute of Technology Faculty of Biomedical Engineering marks a significant advancement toward rapid proteome analysis, with far-reaching implications for both fundamental research and disease diagnosis.
A 2023 Nature opinion article identified the seven most important emerging technologies to watch — with single-molecule protein sequencing taking the top spot. The article highlighted the pivotal contributions of Prof. Meller’s research group in advancing key technologies toward the commercialization of protein sequencing systems, paving the way for a true revolution in biological and medical research.

Since proteins are the body’s fundamental building blocks, their rapid and precise identification is crucial for understanding human health and disease. In a recent paper published in Nature Nanotechnology, Prof. Meller reports a breakthrough that brings this vision significantly closer to reality.
Prof. Meller, who led the research together with postdoctoral fellow Dr. Neeraj Soni, is a faculty member in the Faculty of Biomedical Engineering and the Faculty of Biology, and a member of the Russell Berrie Nanotechnology Institute (RBNI) at the Technion. The research was conducted in collaboration with scientists from the University of Illinois and Rice University.

The technology presented in the Nature Nanotechnology paper applies Prof. Meller’s unique approach – mapping biological molecules using synthetic nanometer-scale pores manufactured with advanced nanotechnologies – to identify the “fingerprint” of whole proteins.
Existing methods can already detect proteins passing through nanopores, but identifying individual proteins requires the use of complex molecular motors to slow down protein motion or designing antibodies for each and every protein – processes that limit their sensitivity and increase their cost of analysis.
Prof. Meller’s technology streamlines protein analysis by allowing precise control of protein movement through a nanopore using a stick–slip mechanism – alternating between rapid adhesion and sliding. As each protein traverses the pore, the system records the ionic current, generating a distinctive electrical “fingerprint” unique to that molecule. A machine learning (ML) model then decodes these current signatures to identify proteins at speeds several orders of magnitude faster than current methods. Each protein’s passage takes only a fraction of a second, enabling near real-time identification. Given that a single human cell contains millions of proteins, this approach represents a potential breakthrough in comprehensive proteome analysis.
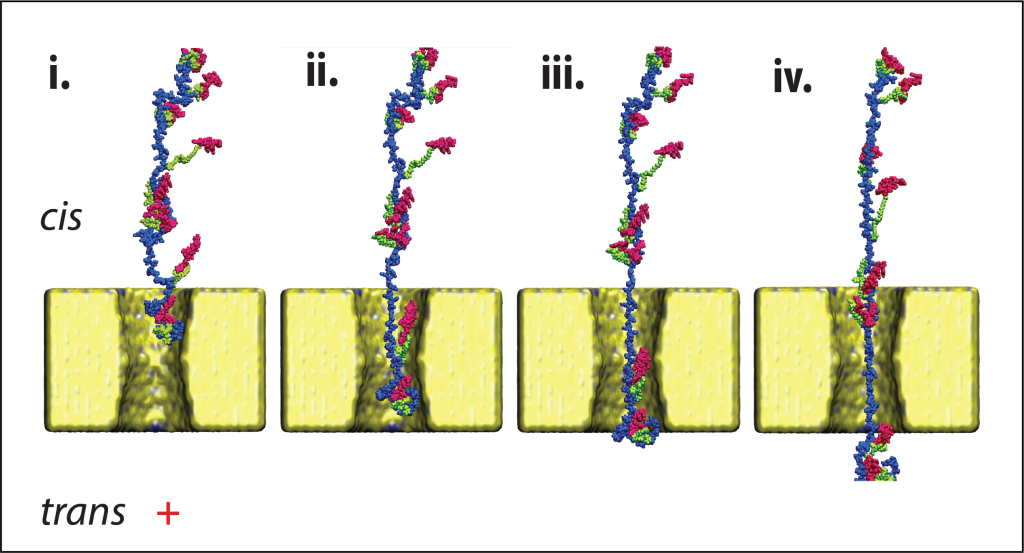
The study focused on the amino acid cysteine, which is essential for many physiological functions in the cell and the body. Since about 97% of human proteins contain cysteine or cysteine residues, this approach applies to nearly the entire proteome. Furthermore, the technology is not limited to a specific amino acid, and the team is already working to expand it to include many others, including those that undergo post-translational modifications (PTMs).
The new method has a wide range of potential clinical applications, including cancer diagnosis and personalized treatment based on simple tests such as blood samples. Scientifically, it is expected to advance protein research and broaden its applicability to a wide variety of proteins. According to Prof. Meller, the long-term vision is to develop an integrated point-of-care platform for rapid protein diagnostics in hospitals, laboratories, and clinical research.
The research was supported by the European Research Council (ERC Advanced Grant) under the Horizon 2020 program.
Link to the paper in Nature Nanotechnology here



