Smart Integration of Single-Cell Sequencing – Toward Early and Accurate Disease Diagnosis
Dr. Dvir Aran from the Technion Faculty of Biology has developed a computational method that extracts important information from single-cell RNA sequencing
Characterizing individual cells is a major technological challenge in biology and medicine, particularly in disease diagnosis. Such characterization enables identification of cellular disruptions that indicate the presence of the disease at an early stage. One way to characterize a single cell is RNA sequencing (scRNA-seq). Although this type of sequencing provides a high-resolution “snapshot” of the individual cell, the resulting data is complex and noisy, making it difficult to distill insights needed for identifying disruptions and diseases.
A pioneering platform developed by Dr Dvir Aran from the Technion Faculty of Biology provides high-quality integration and analysis of data obtained from RNA sequencing of multiple single cells. The platform, called CellMentor, is presented in the journal Nature Communications. In addition to Dr. Aran, the research was co-authored by graduate students Or Hevdeli, who completed her master’s degree in the Aran lab, and Kate Petrenko, who earned her master’s degree under Dr. Aran’s supervision and continued to a Ph.D. in the Rappaport Faculty of Medicine.

CellMentor is a machine-learning method based on a new approach the researchers call “cell-type aware dimensionality reduction.” The tool demonstrates excellent performance:
- Successful analysis in both complex simulations and real-world data
- Effective analysis across sequencing of various cell types, such as blood, pancreatic, or melanoma cells
- Strong performance even in characterizing cell types that are extremely rare in a sample (around 1%)
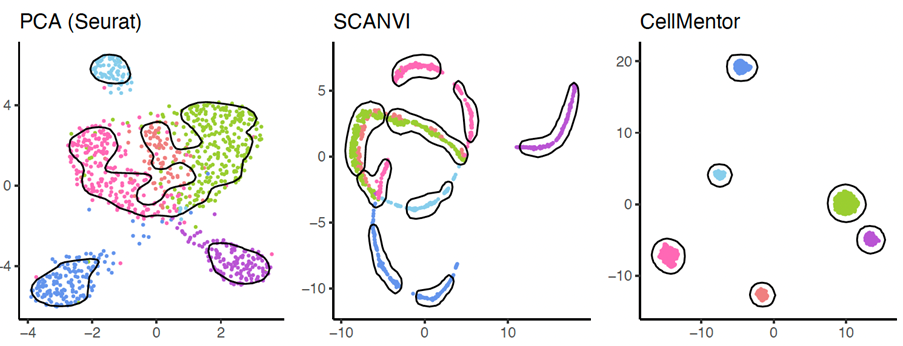
In the illustration: CellMentor’s excellent performance (right) in analyzing information obtained from RNA sequencing. The clear separation between different cell types is visible. In the two other columns: the performance of two popular methods that fail to handle the data and cannot present the cells as distinct units.
According to Dr. Aran, “This tool is effective not only in theory. In a separate study in our lab, we used CellMentor to analyze cells from a solid tumor (neuroblastoma) and discovered a mechanism that could not have been detected using standard methods. In the Nature Communications paper, we present a new and efficient platform for integrative analysis of scRNA-seq data from diverse sources. This platform is already aiding in the integration of multiple datasets, deepening our understanding of complex tissues, and advancing disease research based on cellular patterns.”
The research was supported by the Israel Science Foundation and the Azrieli Foundation.
To read the full article, click here