Disordered is the New Functional: How Transcription Factors Search the Genome
Technion researchers uncover how disordered regions enable transcription factors to search DNA efficiently and selectively, providing direct mechanistic insight into gene regulation
Transcription factors are proteins that control which genes are turned on or off by binding to specific sites on DNA. Traditionally, this ability was thought to rely on the protein’s three-dimensional structure, which fits its target sequence like a key in a lock. Yet how transcription factors locate these sites among millions of similar sequences has long remained a mystery. A new study from the Technion – Israel Institute of Technology Faculty of Biology, recently published in Nature Communications, offers a surprising answer: it is the flexible, unstructured regions of the protein that enable it to find the right target. Using optical tweezers, an advanced technique that employs focused laser beams to manipulate and measure single molecules, the researchers showed that these disordered regions allow the yeast transcription factor Msn2 to loosely attach to DNA and slide along it until it finds its precise target sequence.
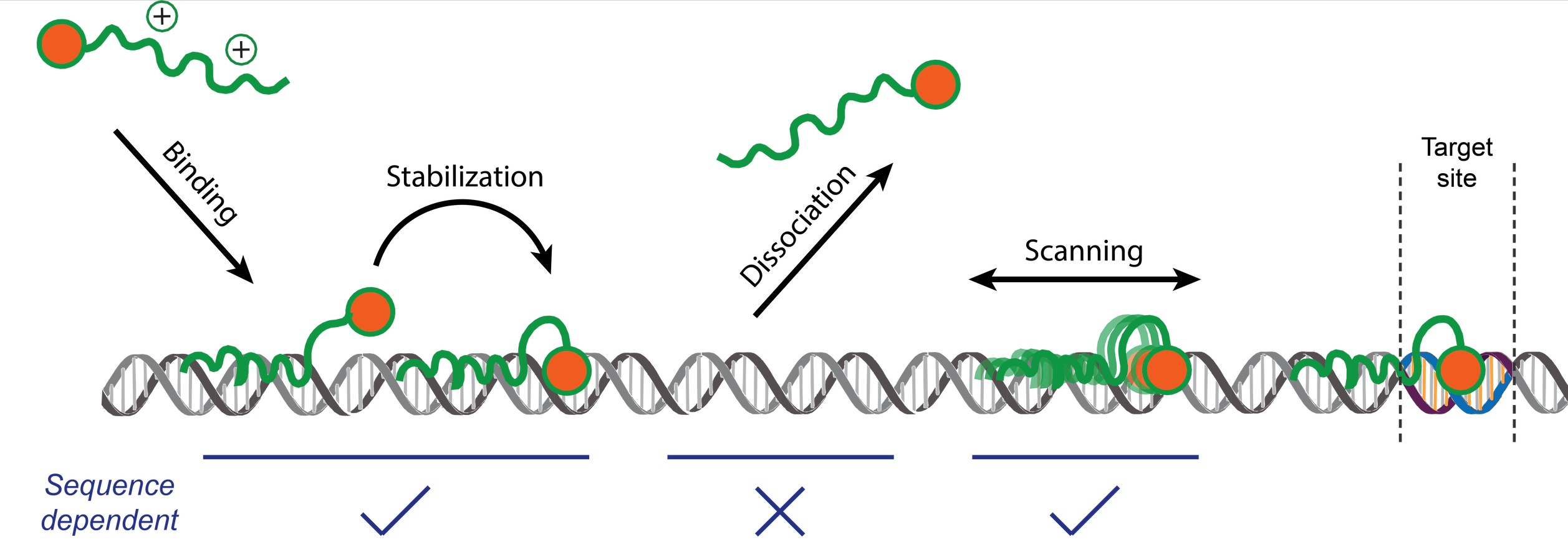
The study, led by Dr. Nir Strugo from Professor Ariel Kaplan’s lab, provides the first direct mechanistic evidence that disordered protein regions can enhance both the efficiency and specificity of transcription factor search, offering new insight into the physical principles of gene regulation.

The research was supported by the Israel Science Foundation.
To read the full article, click here



