 They hope that in the future this will enable the translation of the genome’s “operating system”; they are working in the new field of synthetic biology and believe that this will be “the high tech of bio tech”
They hope that in the future this will enable the translation of the genome’s “operating system”; they are working in the new field of synthetic biology and believe that this will be “the high tech of bio tech”
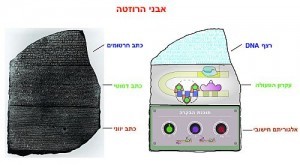
The objective of the Technion researchers is to decode the “software” that controls the process and use this knowledge to develop medical applications. “In order to do this, we intend to create a ‘Rosetta Stone’ for the gene regulatory code (the original Rosetta Stone is a granodiorite stele that had the same ancient text inscribed on it in three different languages, as a result of which archaeologists were able to decipher Egyptian hieroglyphics),” says Dr. Amit. “This tool will be used to ‘hack’ the control program of real organisms and consequently allow us to ‘write’ new programs – which do not exist in nature – for medical purposes, environmental applications, etc. Synthetic biology is a new branch of life science, which takes a constructive/building approach. It attempts to use biological components to construct new biological systems that do not exist in nature. It forces us to really examine our understanding by requiring us to use what we think we understand in order to create biological functions. It allows us to ask why evolution “locked onto” specific patterns, to imagine and create new biological functions and forces us to work in a multidisciplinary fashion.”
The approach of researchers in synthetic biology is based on using characteristic genomic components and arranging them together (or “wiring” them to each other) in new architectures. In the next stage they develop patterns based on thermodynamic models, and in the end, they analyze the output using their model. By doing this, they can draw basic programming principles that permit them to translate the architecture and the sequence into computer algorithms. “If we succeed in writing a sequence that predicts our output based on computerized rules that we found in the ‘Rosetta Stone’ – we can then use this ‘key’ to decipher certain sequences that appear in the genome,” says Dr. Amit.
In the paper appearing in Cell, the Technion researchers show that they can use this approach to develop a new understanding of enhancers among bacteria. These sequences are common to all living creatures and may be thought of as modular objects that can combine “input” or signals. Because bacterial enhancers have a simpler architecture and at times it is easier to characterize them, the Technion researchers chose to focus on them first. The researches demonstrated the possibility of building new bacterial enhancer programs that will lead to a physical model of the control program, or to the “machine code”. The researchers note that the type of computerization that takes place in this context is reminiscent of analogue computing processes more than digital ones.
“This Rosetta Stone, in the bacterial context, has enabled us to formulate a new understanding, or qualitative model, for many examples of bacterial enhancers in nature, most of which have never been analyzed,” stresses Dr. Amit. “Now we can understand, at least partially, many natural programs that have not yet been decoded by simply reading the DNA sequence.”

 They hope that in the future this will enable the translation of the genome’s “operating system”; they are working in the new field of synthetic biology and believe that this will be “the high tech of bio tech”
They hope that in the future this will enable the translation of the genome’s “operating system”; they are working in the new field of synthetic biology and believe that this will be “the high tech of bio tech”