Primordial Reproduction
Technion researchers and international colleagues hypothesize that the cellular mechanisms enabling the fusion of sperm cells and egg cells originated 3 billion years ago
The process of sexual reproduction in plants and animals, as it is familiar to humans, requires the fusion between egg cells and sperm cells. The emergence of sexual reproduction is usually dated back about 1-2 billion years ago. But Technion – Israel Institute of Technology researchers, in collaboration with expert teams abroad, now speculate that the mechanisms that allow for this fusion event to occur appeared as early as 3 billion years ago.
The research, published in Nature Communications, was carried out by Professor Beni Podbilewicz and doctoral student Xiaohui Li from the Technion’s Faculty of Biology, along with researchers from Uruguay, Switzerland, Sweden, France, Great Britain, and Argentina.
The fusion of sperm and egg marks the climax of fertilization – and the onset of embryonic development. Since both cells contain exactly half of the genetic information needed for the offspring, aberrant fusions of multiple sperm to one egg will have abnormal development. The process is therefore tightly regulated. Specialized proteins called ‘fusogens’ must be present at the precise time and place to allow the egg and the sperm to merge into one.
The Podbilewicz Laboratory studies fusogens in several organisms, and first identified and characterized two such proteins in the nematode C. elegans (EFF-1 and AFF-1). These proteins are involved in organ development, but not in fertilization. Surprisingly, structural analysis revealed that these proteins have a very similar three-dimensional structure to another fusogen involved in fertilization in plants (GCS1/HAP2). This family of similarly structured fusion proteins has been named Fusexins and includes representatives in plants, animals, viruses, unicellular algae, and parasites.
In order to expand upon and to characterize the origin and evolution of the Fusexin family, research collaborator David Moi and a team of researchers from Argentina and Switzerland conducted a bioinformatic search on genetic sequences sampled from different environments. After screening samples from soil, saline lakes, freshwater, and marine sediments, they discovered 96 sequences belonging to archaea, which showed some similarities with known fusion proteins. The sequences were named fusexin1 (Fsx1), and an expert team led by Pablo S. Aguilar, Hector Romero, and Martin Graña confirmed that they belong to archaea species from lineages estimated to originate 3 billion years ago. However, it remained unclear whether the protein that Fsx1 encodes looks similar to members of the fusexin family and whether it is truly capable of mediating cell-to-cell fusion.

The research group from the Technion, from right to leftR-L: Prof. Benjamin Podbilewicz, Katerina Flyak, Clari Valansi, Xiaohui Li, and Dr. Nicolas Brukman
To determine the structure of an Fsx1 protein, Shunsuke Nishio of the Karolinska Institute in Sweden used crystallographic methods to decipher the three-dimensional conformation of the Fsx1 protein. Nishio showed that the Fsx1 protein contains three structural domains very similar to known Fusexin members and is arranged in a three-piece complex – known as a trimer – as do other known fusogens. Surprisingly, Fsx1 has an additional fourth domain not found in any known Fusexin member. Professor Luca Jovine, leading the crystallographic structure analysis, also used novel machine-learning software (AlphaFold2) to determine the structure of the Fsx1 protein.
To prove that the protein Fsx1 carries a fusogen role, doctoral student Xiaohui Li from the Podbilewicz Laboratory conducted an experiment in which she expressed the Fsx1 protein in a cell culture derived from mammals, which typically do not fuse. In collaboration with lab manager Clari Valansi and lab members Dr. Nicolas Brukman and Kateryna Flyak, Li showed that Fsx1 from archaea does induce the fusion of these mammalian cells that diverged 1-2 billion years ago.
Known Fusexin proteins in viruses serve to mediate viral entry into the host cell (as is the case for coronavirus), while in eukaryotes (cells with nuclei) – plants, nematodes, and protists – they play roles in organ sculpting, neuronal repair, and sex. But who came first? Was a fusogen used for sexual reproduction snatched by a virus, or was a viral protein used for infection adopted by plants? Since Archaea predate the origin of eukaryotes, the study by Moi, Nishio, Li, and others raises a possible third scenario: all fusexins originate in archaea, from which the lineage split into a variety of functions, from viral infection to sperm and egg fusion – a billion years before sexual reproduction.
An important next step will be to study what Fsx1 proteins are doing in nature. Do they fuse archaeal cells like their plant and animal fusexins counterparts fuse gametes (e.g., eggs and sperm) to promote a sex-like DNA exchange? Parallel studies will also be needed to understand the evolutionary history connecting the Fsx1 protein and GCS1/HAP2 in order to establish their origin. Archaeal fusexins and other still undiscovered fusogens might help us to understand how cells evolved from apparently simple forms sharing discrete pieces of DNA between them to today’s complex life forms undergoing sexual reproduction. Thus, the discovery that ancient creatures like archaea can also contain fusexin proteins now raises the intriguing possibility whereby the Fsx1 protein is the ancestral version from which viral, plant, and animal fusogens derived.
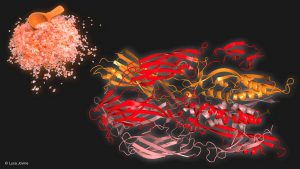
An illustration by Prof. Luca Jovine of the Karolinska Institute shows the Fsx1 protein structure (on the right) deciphered by the researchers, belonging to an archaeon from a hyper-saline environment (represented by salt on the left).
The study was supported by the European Union’s Horizon 2020 program, the Marie Curie Grant, the Israel Science Foundation, and others. Some sample analysis was conducted at the European Synchrotron (ESRF) facility.
For the full article in Nature Communications click here.



