The Computerized Pathologist
New technology developed at the Technion is expected to improve personalized care in cancer treatment
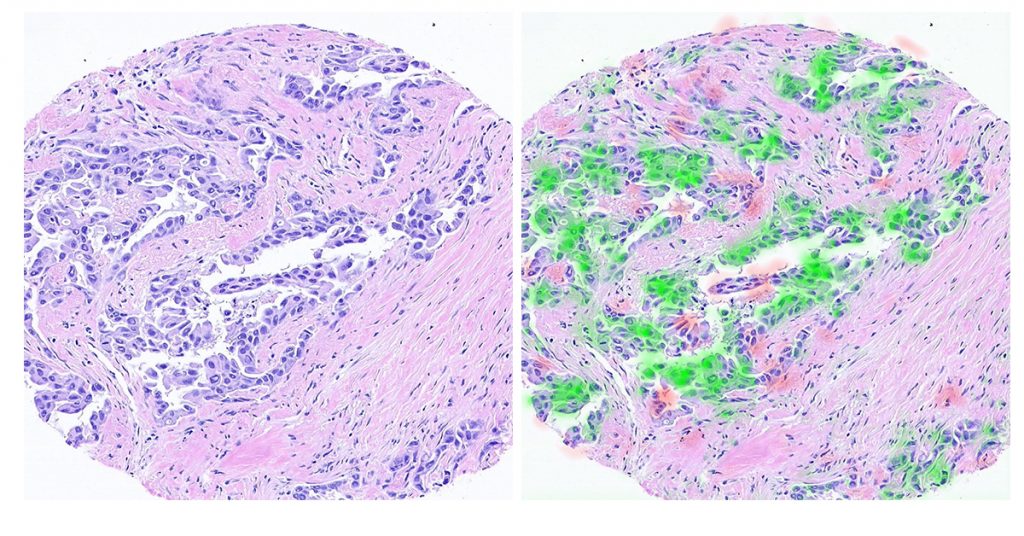
The original scan (left) and the areas where information was extracted (in red and green, right) using the technology developed at the Technion
Credit: Technion Spokesperson Department
Technion researchers have developed a deep learning-based method for mapping critical receptors on cancer cells. Using digital images of biopsies taken from breast cancer patients, the new technology is expected to significantly improve personalized cancer treatments. Published in the prestigious JAMA journal, the research was conducted by doctoral students Gil Shamai and Ron Slossberg and Professor Ron Kimmel of the Technion Faculty of Computer Science, in collaboration with Dr. Yoav Binenbaum of Ichilov Hospital and Professor Ziv Gil of Rambam Medical Center.

Prof. Ron Kimmel
The technology extracts molecular information from biopsy images that underwent hematoxylin and eosin (H&E) staining. H&E is a common dye used to test tissue taken in a biopsy. The staining allows the pathologist to identify the type of cancer and its severity in the tissue under the microscope. But staining alone does not allow the identification of characteristics that are crucial in determining the appropriate treatment. These include the molecular profile of the tumor, its biological pathways, the genetic code of the cancer cells, and the common receptors on the cell membrane. The mapping of receptors on the cell membrane is particularly relevant to personalized medicine, since it enables matching cancer patients with the treatment that will block the receptors and inhibit the development of the tumor.
The Technion researchers’ conceptual innovation is in extracting molecular information from the cell shape and the environment (the morphology of the tissue) as reflected in the H&E scans. According to Mr. Shamai and Prof. Kimmel, “Pathologists we spoke to said it was an impossible task. A human pathologist cannot infer the tumor features from its shape because of the sheer number of variables. The good news is that artificial intelligence technologies, and especially deep learning, are capable of doing so. The computer, unlike even the most skilled pathologist, can characterize the cancer with a complex analysis of its morphology.”

Doctoral Student Gil Shamai
With the help of image processing and artificial intelligence tools, the researchers showed, for the first time, the ability to predict the molecular profile of cells from tumor morphology, that is, only from looking at the tissue as it appears on standard H&E scans.
“We succeeded in identifying the ‘signature’ that the cancer leaves in the tissue,” Mr. Shamai explains. “It’s a morphological signature (formative) that, through our technology, we are able to glean essential information. It is important to note that deep learning systems require a huge amount of information and obtaining the kind of information required is not easy. To that end, we have written software code to scan network sources and automatically download thousands of biopsy samples and the relevant medical information approved for research.”
The study examined more than 20,000 scans from 5,356 breast cancer patients. Using the new technology, the researchers were able to map estrogen and progesterone receptors, among other molecular biomarkers, from the scans alone and based on cell morphology. The study focused on breast cancer, but the researchers make it clear that this is a feasibility study relevant to all cancers.

Doctoral Student Ron Slossberg
According to Prof. Kimmel, “We have succeeded in showing that cancer has a unique signature in tissue morphology and that computerized mapping of this morphology can give us tremendously relevant information on tumor characteristics. In the first phase, we believe it will be a tool to help doctors make decisions and will later be developed as a real clinical tool.”
The research was supported by the Ministry of Science and Technology, the National Science Foundation, the Lorry Lokey Interdisciplinary Center for Life Sciences and Engineering, and Schmidt Futures.
For the full article on JAMA click here.


