Efficient Protein Synthesis
Researchers from the Technion Faculty of Biology have discovered a sophisticated, autoregulatory mechanism that makes protein synthesis in the human body more efficient
The researchers say damage to this mechanism may be linked to many illnesses
Researchers from the Technion Faculty of Biology have discovered a sophisticated regulatory mechanism that makes protein synthesis in the human body more efficient. The researchers say damage to this mechanism may be linked to many illnesses, and that their findings are likely to lead to a better understanding and improved treatment of biological disruptions. The findings of the study led by Professor Yoav Arava and doctoral student Ofri Levi were recently published in PLoS Biology.
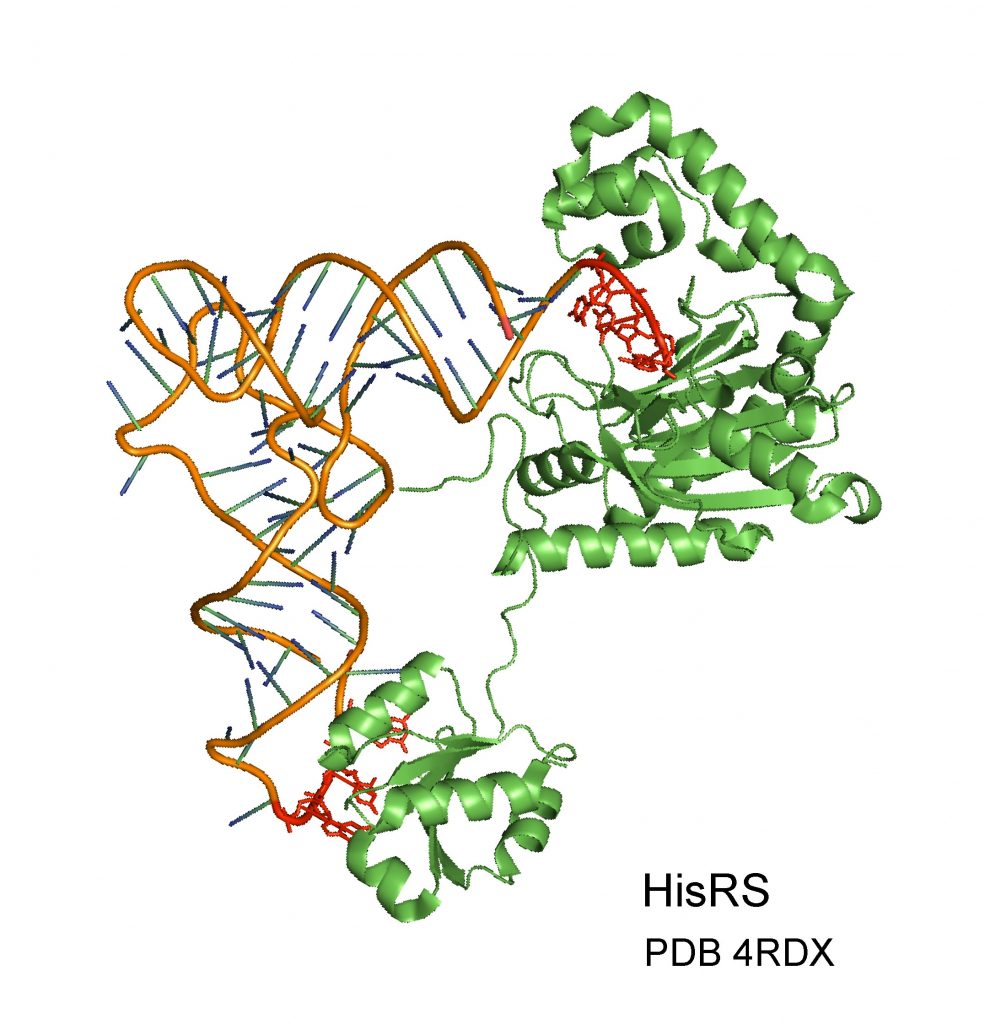
The Aminoacyl-tRNA synthetases enzyme (green) identifies a tRNA molecule (brown) and binds it to an amino acid. The molecule is correctly identified according to the regions marked in red. The Technion researchers discovered that the enzyme also knows how to identify other types of RNA molecules (mRNA). Moreover, correct identification of the mRNA depends on regions that are very similar to the identification regions along the tRNA (marked in red).
Proteins, a major component of the human body, are synthesized by a cellular factory known as the ribosome. For protein synthesis, the ribosome uses two main components: mRNA, which contains the necessary code for protein synthesis, and tRNA, which carries the amino acids, the protein’s building blocks.
The research conducted at the Technion focused on a unique family of enzymes –Aminoacyl-tRNA synthetases (aaRSs) – that play an important role in properly binding the amino acid molecule to the corresponding tRNA. The precision and efficiency of these enzymes are critical to the quality of the raw materials reaching the ribosome and hence to the quality of the future protein. While research groups around the world have been investigating the role of these enzymes for decades, the Technion researchers discovered another, previously unknown function.

Doctoral student Ofri Levi
Prof. Arava’s research group discovered that in addition to binding the amino acids to their matching tRNA, these enzymes also regulate their prevalence in accordance with the amount of available amino acids. When amino acids are abundant and large quantity of enzymes are required, the enzymes activate an autoregulatory mechanism that increases their production. The opposite occurs when a smaller quantity of enzymes is needed. Furthermore, the researchers deciphered the molecular mechanism that executes this action: the enzyme binds its own mRNA in a region that mimics the binding site on its natural tRNA target.
Even though in the current research the Technion researchers focused on a single enzyme from the Aminoacyl-tRNA synthetases family, they believe that this autoregulatory mechanism exists in all twenty enzymes belonging to this family. This intelligent mechanism enables the molecule to regulate its amount in accordance with the cell’s needs and probably developed in an early stage of evolution.
Due to the critical role of Aminoacyl-tRNA synthetases in determining the quality of the raw materials in the ribosome, any disruption in their activity is likely to lead to the synthesis of harmful proteins. Indeed, mutations of these enzymes are involved in many human illnesses, in particular those related to the nervous system such as amyotrophic lateral sclerosis (ALS) and Charcot-Marie-Tooth (CMT). Hence, this research may lead to a better understanding of illnesses and to the development of new ways to treat them.
The research is funded by the Israel Science Foundation (ISF), the US-Israel Binational Science Foundation (BSF) and the Russell Berrie Nanotechnology Institute (RBNI) at the Technion.

Prof. Yoav Arava
Prof. Yoav Arava completed his bachelor’s degree in the Faculty of Agriculture in Rehovot, his master’s and doctorate in the Faculty of Biochemistry at the Weizmann Institute of Science and his post-doctorate in the Department of Biochemistry at Stanford University. He has been the recipient of many awards, including the Yanai Prize for Excellence in Academic Education (2012) and the Mani Award for Excellence in Teaching (2009). Prof. Arava’s laboratory focuses on proteins that bind to RNA molecules and on the biochemical and molecular attributes of their functioning. “We use two main experimental systems: yeast cells (used for the research described here) and nerve cells,” he said. “By generating genetic changes in these systems we attempt to understand the communication between proteins and RNA and its contribution to the physiology of the cell. The current paper is a classic example of the work in our laboratory: we identified a link between proteins and particular RNA molecules, and by generating genetic changes in the protein or the RNA we were able to expose a new physiological function and its underlying mechanism.”
For the article in PLOS Biology click here


