Molecular Fingerprint of Animal Kingdom
Redefining part of 300 year-old classification system for grouping members of the animal kingdom
New “molecular fingerprint” of animal kingdom emerges from gene regulation survey
An international team of biologists has identified the molecular signature of the animal kingdom, providing genetic evidence for an animal classification that has been used for nearly 300 years. Their research, published this week in the journal Nature, offers a historic dataset for the field, serving developmental biologists, evolutionary biologists, and computational biologists alike.
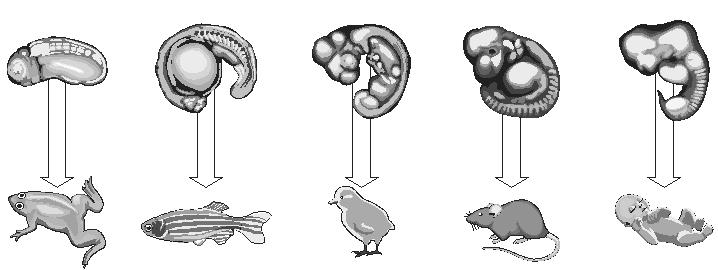
The study was led by Professor Itai Yanai of the Technion-Israel Institute of Technology Department of Biology, in cooperation with research teams in Australia, Germany, the US, and Israel. The research team investigated an extremely diverse set of animal species, applying an extremely powerful technique called CEL-Seq, developed in 2012 by Dr. Tamar Hashimshony in the Yanai lab. CEL-Seq allows for the monitoring of the activity of all genes in individual cells, and the team used it to analyze gene regulation in 70 embryos in each of ten species.
The researchers found a striking pattern of universality across the species. Between phases of similar genes turned ‘on’ at the beginning and the end of development, a mid-developmental transition was discovered. This newly discovered gene regulatory pattern explains how the differences among animals develop and evolve, which allows biology to now have molecular means to define the specific properties of groups of species.
Their work further defines a category of animal life under-defined since 1735 when Swedish botanist Carl Linnaeus, recognized as the father of the biological classification of organisms, proposed a two-name classification system for the world’s plants and also animals classified animals into “families” based on similarities and differences in body “plans.” The work sheds new light on how, at the molecular and genetic levels, animals of different body designs (whether they have a true spinal column (mammals) or just a nerve cord (chordates) have evolved to be different and why.
Nearly eight million different species of animals are thought to inhabit the planet, covering a striking exuberance of diversity. For example, animals span five orders of magnitude of adult body sizes. Prof. Yanai’s team began this research by asking what is common to all animals. To tackle this question, they chose ten of the most different animals one could choose: a fish, a worm, a fly, a water bear, a sponge, and five others, each of a different phylum (a term coined by German naturalist Ernst Haeckel in the 19th century to describe a group of animals with the same body plan). About 35 phyla are typically recognized, however it remains controversial with contention over whether this is a meaningful classification and, if so, what attributes are the same, or different across all animals.
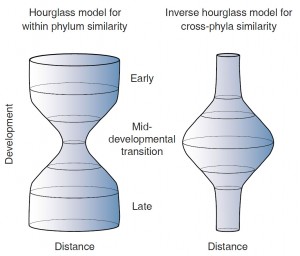
Inverse hourglass model for the origin of phyla compared with the hourglass model for within phylum evolution
“We selected species representing ten different animal phyla,” said Prof. Yanai. “For each phyla we determined the gene expression profile of all genes from the development of the fertilized egg to the free-living larvae. We found a surprising pattern of gene expression conservation in all species occurring at a pivotal, transitional period in development.”
By studying the molecular programs of development in ten very different animals, the researchers found that all of the animals they studied express two distinct “modules” of genetic expression. (A module is a set of genes – similar across the organisms – that are turned ‘on.’) During the transition between the modules, mechanisms of cell signaling and regulation occur.
With this new knowledge, the researchers proposed a definition for phylum as “a set of species sharing the same signals and transcription factor networks during the mid-developmental transition.” In other words, they clarified the definition by suggesting that those organisms sharing a phylum, formerly by virtue of body design alone, also share a unique and similar genetic and molecular transition that other species do not.
To demonstrate their proposal, the researchers developed an “hourglass model” that captures gene expression differences between species. The inverse hourglass model shows the origin of phyla compared with the hourglass model that demonstrates “within phylum evolution.”
Embryonic development is called the “phylotypic” stage. This is when the embryo begins to assume recognizable features typical of vertebrates. The phylotypic stage represents a general layout on which specialized features—such as the turtle’s shell, the pig’s snout, or your large brain—can be mounted later in development.
The researchers proposed that during the phyletic transition period, properties specific to each phylum are genetically encoded. Their emerging dataset, they said, will be useful in studying the hallmarks of animal body plan formation from the embryonic stage.
As with many scientific discoveries, the researchers suggest that their work “raises more questions than it answers.” For example, “what molecular pathways underlie phyletic transition in each phylum? Why are the phyletic-transition mechanisms so relatively susceptible to change? Is the coupling of the conserved modules universal to all multicellular life?”
“The transition we identified may be a hallmark of development only in animals,” the researchers concluded. “Or, future work may show that this is a general characteristic of development in all multicellular life.”